Importance of Variables According to the Singh (1981) Criterion
singh.RdA function to calculate the Singh (1981) criterion for importance of variables based on the squared generalized Mahalanobis distance. $$S_{.j} = \sum_{i=1}^{n-1} \sum_{i'>i}^{n} (x_{ij} - x_{i'j}) * (\bold{x}_i - \bold{x}_{i'})' * \bold{\Sigma}_{j}^{-1} $$
# S3 method for default singh(data, cov, inverted = FALSE) # S3 method for singh plot(x, ...)
Arguments
| data | a data frame or matrix of data (n x p). |
|---|---|
| cov | a variance-covariance matrix (p x p). |
| inverted | logical. If |
| x | an object of class |
| ... | further graphical arguments. |
Value
singh returns a matrix containing the Singh statistic, the
importance proportion and the cummulative proprtion of each
variable (column) in data.
References
Singh, D. (1981) The relative importance of characters affecting genetic divergence. Indian Journal Genetics & Plant Breeding, 41:237-245.
Author
Anderson Rodrigo da Silva <anderson.agro@hotmail.com>
See also
Examples
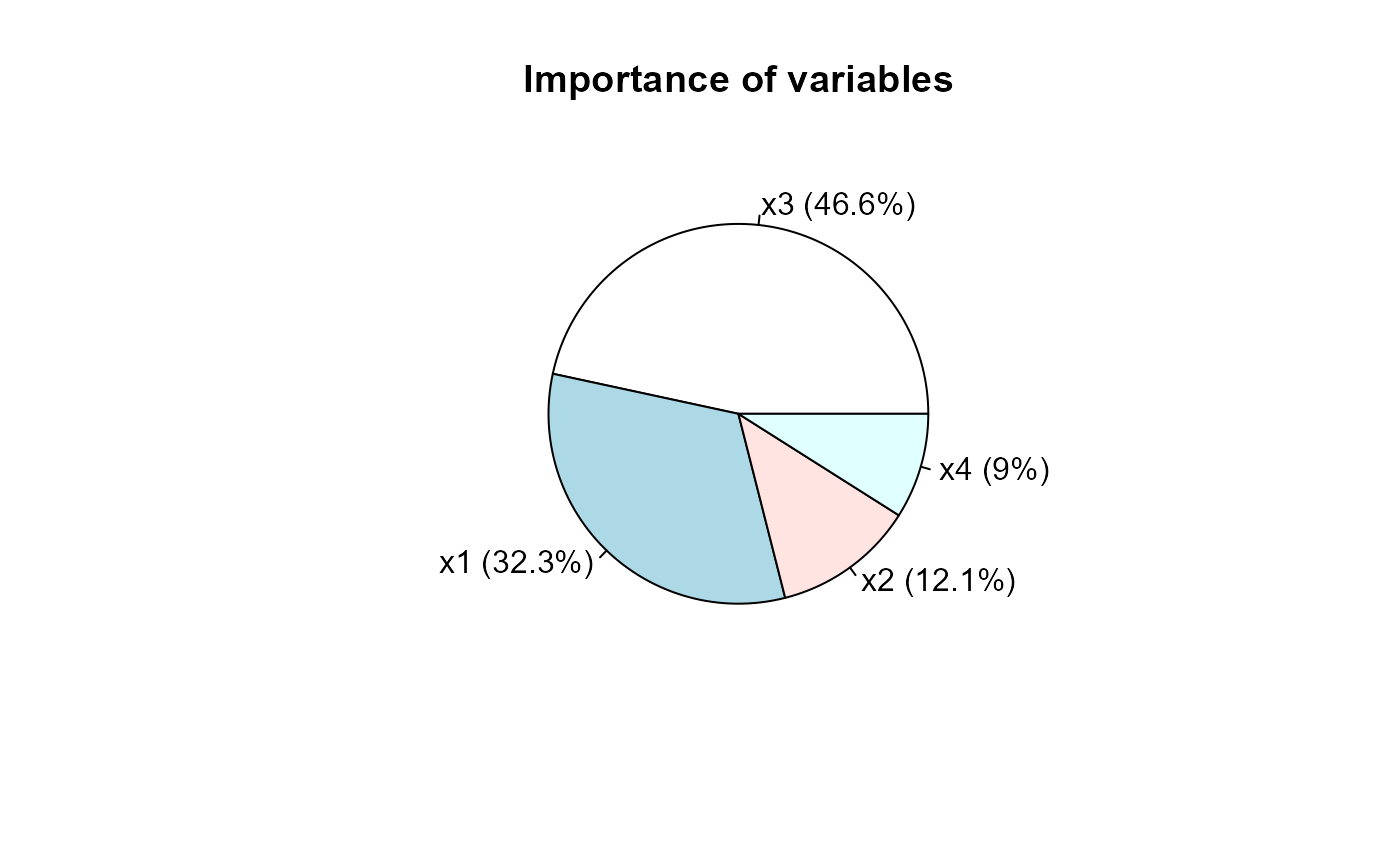
# Manly (2004, p.65-66) x1 <- c(131.37, 132.37, 134.47, 135.50, 136.17) x2 <- c(133.60, 132.70, 133.80, 132.30, 130.33) x3 <- c(99.17, 99.07, 96.03, 94.53, 93.50) x4 <- c(50.53, 50.23, 50.57, 51.97, 51.37) x <- cbind(x1, x2, x3, x4) Cov <- matrix(c(21.112,0.038,0.078,2.01, 0.038,23.486,5.2,2.844, 0.078,5.2,24.18,1.134, 2.01,2.844,1.134,10.154), 4, 4) (s <- singh(x, Cov))#> x3 x1 x2 x4 #> Singh statistic 5.4361584 3.7721798 1.4059118 1.04694241 #> Proportion 0.4661752 0.3234815 0.1205633 0.08978005 #> Cumulative proportion 0.4661752 0.7896567 0.9102200 1.00000000 #> attr(,"class") #> [1] "singh"# End (not run)